Hierarchical Bayesian Analysis of Differential Expression and ALternative Splicing (HBA-DEALS)
HBA-DEALS simultaneously characterizes differential expression and splicing in cohorts.
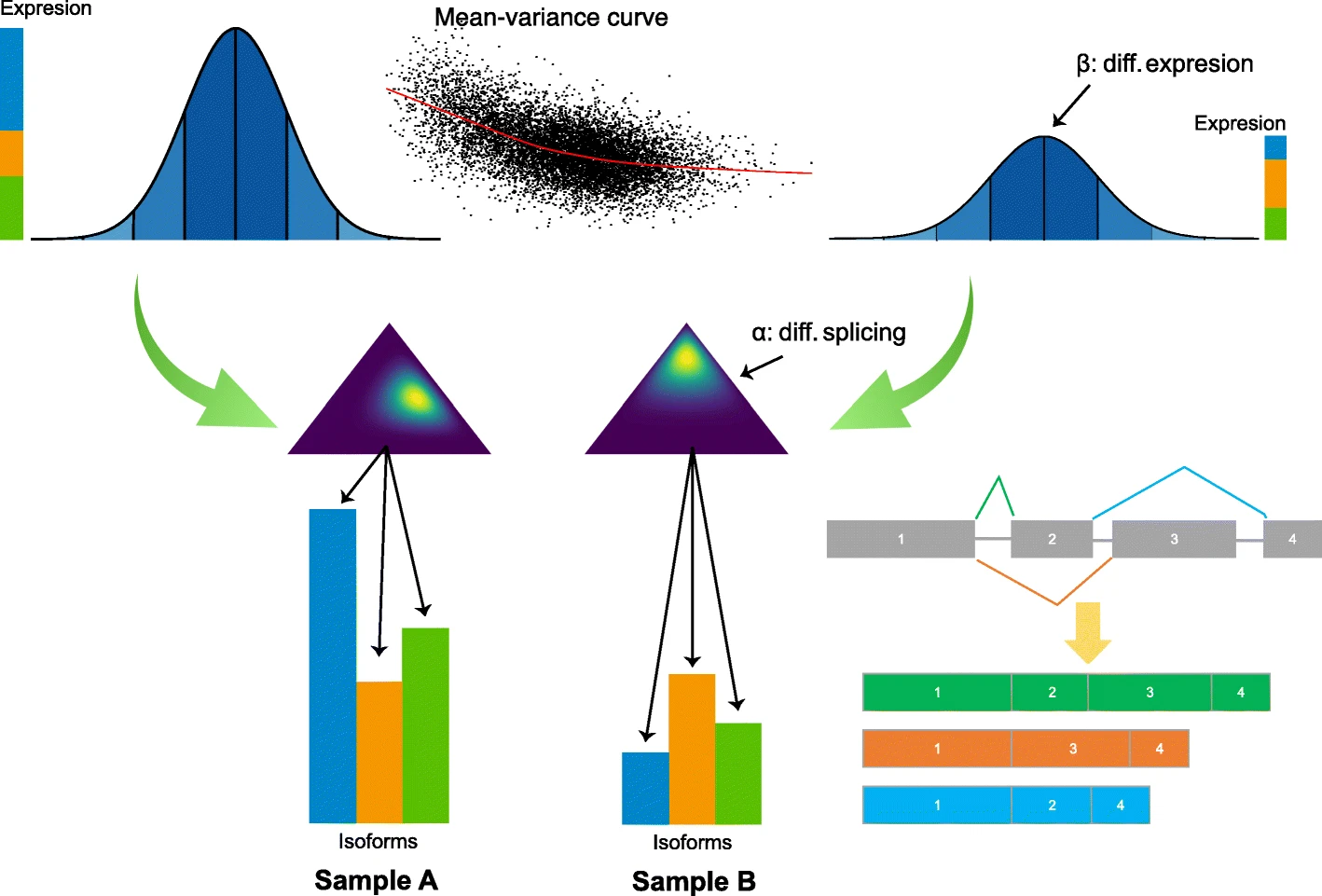
HBA-DEALS is based on a hierarchical Bayesian model of the absolute expression levels of the gene and its isoforms. HBA-DEALS assumes that the data are available from n RNA-seq samples and that the sequence reads have been mapped to isoforms. The n samples are divided into two cohorts n1 and n2 (e.g., cases and controls). The output of any isoform quantification tool, including but not limited to Salmon, RSEM, Kallisto, and StringTie, can be used as the input for HBA-DEALS.
This documentation explains how to setup and run HBA-DEALS.